|
Main Menu |
|||
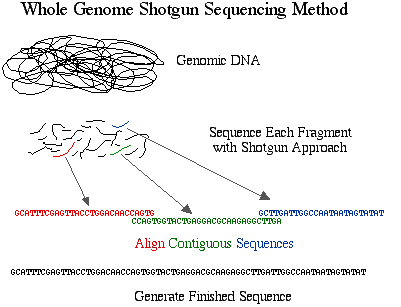
Celera: A Unique Approach to Genome SequencingIn 1981, Gene Myers, a current professor of Computer Science at Berkeley graduated with a PhD in computer sciences from the University of Colorado and joined the faculty at the University of Arizona. While working at the University of Arizona, Myers worked with a geneticist named Jim Weber from Wisconsin in the field of bioinformatics. They came up with a revolutionary approach of large-scale sequencing projects such as that of the Human Genome Project. Their approach was called Whole Genome Shotgun Sequencing. Rather than the traditional method of separating the genome into BACs and sequencing each individual BAC via BAC-end Shotgun Sequencing, Myers proposed the idea of breaking many copies of the entire genome into different sized pieces and sequencing those pieces in a special way. A huge percentage of the human genome consists of repeated non-coding sequences dispersed throughout the genome. These regions included LINEs (Long Interspersed Nucleotide Elements), and Minisatellites: multiple copies of the same sequence that does not appear to have any valid function. Most geneticists considered them to be difficulties in the sequencing process, for they were unpredictable and made the number of base pairs to sequence increase by a huge factor. Myers, instead, used them to his advantage. He proposed that the entire genome be broken up into pieces of 2000, 10000, and 50000 base pairs long. Unique DNA sequences were sequenced while the ends of repeating non-coding sequences were used as markers to rebuild and connect the sequences together, similar to traditional shotgun sequencing. Thus, the principle behind his method was the breaking up of many copies of the whole genome randomly into pieces, and then parsing them together, using repeating elements such as LINEs as identifying markers.
When this idea was proposed to the publicly funded Human Genome Project, however, it was met with great doubts. Many people doubted the ability of Myers' technique to sequence the genome, for they believed it would be prone to errors. J. Craig Venter, the president of Celera at the time, however saw potential in Myers' proposal and signed him on as the vice-president of Informatics Research to test his approach. One can immediately see, however, that in order for his approach to work, the millions upon millions of sequences generated from the project would have to be put together by their ends and their non-coding repeating sequences. To do this, it would take immense amounts of computing power, which is the exact approach Myers and Celera took. Upon signing on with Celera, Myers and other researchers worked to program a 500,000 line code that would be able to run an algorithm capable of matching up non-repeating sequences paired with repeating sequences with other sets of such sequences. Thus, rather than using divide and conquer to sequence the genome from small plasmids to BACs to the genome, Myers developed an algorithm that would be able to assemble the whole genome by matching ends of sequences up, and using repeating sequences almost as scaffolds to identify sequences relative to each other. For this algorithm to run efficiently, however, it required vast amounts of computing power.
Upon the establishment of the genome project at Celera in 1998, the company purchased and connected 700 CPUs and 70 terabites of hard drive space. This computing system was established to run the initial test of their algorithm code, which was used to sequence the genome of the Drosophilla fruit fly with a 13-fold coverage of the genome successfully in 1999. The most surprising thing about this approach was that it succeeded in coding the algorithm and sequencing the 120 Megabase pair genome of the fruit fly to that extent of completeness in just 11 months. Myers then modified the process so that the Whole Genome Shotgun Sequencing process would make a 5-fold coverage of the human genome, as he believed it would be adequate to provide a complete sequence of the human genome. In addition, Venter purchased 4 supercomputers referred to as the GeneMatcher from a company called Parcel Inc. Parcel Inc, a company that typically produces computers for government agencies such as the NSA, created this machine specifically for matching character strings, such as putting together sequences of DNA like a puzzle. It was composed of 7000 processors arranged to perform over 1000 times faster than any Pentium computer. With this new technology, on September 8, 1999, Celera began its sequencing of the human genome using this approach, and completed the first assembly of the whole human genome in June 17, 2000, only 9 months after the project began. It is obvious that modern computing power played a critical role in the completion of the Human Genome Project. Rather than relying on human resources of different parties in a divide-and-conquer strategy, Celera used computers to assemble the whole genome. Rather than dividing the genome into pieces for different parties to sequence, they broke up the entire genome at random and sequenced them like pieces of a very large puzzle. This puzzle, which normally would have been impossible to solve by traditional means, was pieced together in a short period of 9 months, assembling a 5-fold coverage of the human genome, consisting of 14.8 billion base pairs. Had it not been for Celera's project, the genome would not have been able to be successfully sequenced for a long time. It is perhaps the best example of the possibilities that modern computing power offers in the field of genetic research, for it allowed Myers and his team at Celera to accomplish in 9 months what the international NIH funded Human Genome Project could not complete even in 15 years. |
||