Dissecting thermotolerance mechanisms in Thermothelomyces thermophilus.

Understanding the evolution of marine fungi.
Fungi are predominantly terrestrial organisms, and our understanding of marine fungi remains limited. While many extremophilic and extremotolerant fungi have been isolated from marine environments, there also exist numerous obligate marine fungi across diverse lineages that have diverged from largely terrestrial ancestors. Our research seeks to uncover the strategies these fungi have evolved to meet the unique challenges of marine ecosystems and to understand why distantly related species have repeatedly made this transition back to the sea.
One of our current model systems is Kluyveromyces nonfermentans, a yeast isolated from the deep sea. The Kluyveromyces clade includes both terrestrial species and species from brackish waters and estuaries, offering a framework to explore how K. nonfermentans transitioned into marine habitats and specialized for life in the deep ocean. Through this model, we aim to identify the traits that are unique to K. nonfermentans and determine how these adaptations enable its survival in extreme deep-sea environments.

A cold seep in teh South China Sea (Xudong Wang et al. Geosystems and Geoenvironment, 2022).
Sexual transmission of Botryozyma yeast between Panagrellus nematodes.
Many traits of industrial and biomedical interests arose long ago and are fixed in a given species, distinguishing it from its relatives. Understanding how evolution builds a trait over long timescales is a central goal for evolutionary geneticists. Botryozyma yeast spp. serve as a good system to investigate such question. We have found that this species has an ability, unique in the fungal kingdom, to attach to Panagrellus nematodes, especially their reproductive organs. The yeast can be transmitted between male and female nematodes, rendering it a powerful model for sexually transmitted infections. Current work focuses on the genetics of the unique trait and the impacts of the yeast on the host and vice versa, using both computational and wet lab approaches.
Panagrellus redivivus nematodes’ reproductive organs attached by Botryozyma nematodophila. (A) A female P. redivivus with B. nematodophila on the vulva. (B) A male P. redivivus with B. nematodophila on the spicule. 20X, B. nematodophila was stained with Calcofluor white.
Mapping genotype to phenotype in fungi.
Most fungal genomes are poorly annotated, so we have no idea of the genetic architecture for many fungal traits of industrial and biomedical relevance. In published work, our lab has used variation across wild populations to find the molecular basis of morphology traits in the model fungi Saccharomyces and Neurospora; we have also developed genomic screening approaches to annotate candidate virulence islands in the plant pathogen Zymoseptoria and lipid metabolism genes in the oleaginous yeast Rhodosporidium. Our next project, now in progress, is focused on strain variation in virulence and stress resistance traits in the human pathogen Coccidioides.
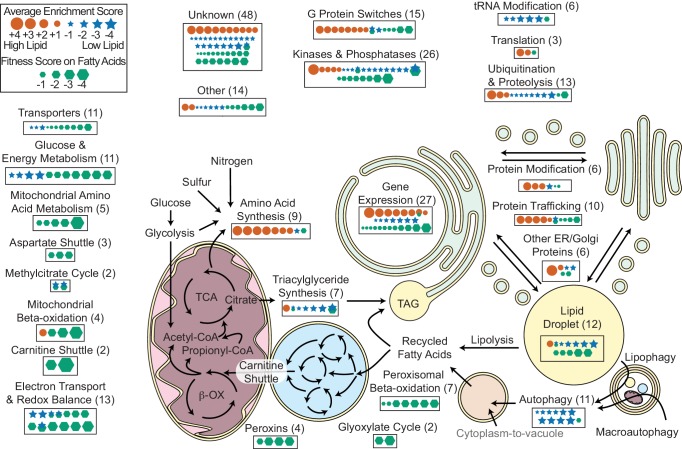
Key metabolic pathways and cellular functions mediating lipid metabolism as identified from fitness scores on fatty acid and enrichment scores from lipid accumulation screens. From Figure 6 of Coradetti et al., 2018
Differences between mouse species in lysosome titer and function.
The lysosome is the protein recycling hub of the cell and also acts to direct cell growth decisions. These functions break down during aging and in many disease states. The prospect of interventions to boost lysosomal function is of abiding interest in biomedicine; for the moment, the literature reporting genetic or pharmacological manipulations toward this end remains in its infancy. We have found a robust lysosomal acidification phenotype in the fibroblasts of a little-studied wild Mediterranean mouse species, Mus spretus, in comparison to cells from commensal M. musculus. This character is accompanied by dampened lysosomal enzyme activity and expression in M. spretus cells, as expected if the well-functioning lysosomes of the latter require no compensation at the regulatory level. We find tight conservation of these traits within species against the backdrop of marked differences between species, consistent with a history of natural selection. These results matter because they attest to a program of extra-avid lysosomal function in M. spretus, of the kind associated with healthy aging and disease resistance that the field has sought in the laboratory setting for decades. Our next step is to find the underlying genes, for which omics analyses are ongoing.

M. spretus (B) shows more green fluorescence signals, indicating more acidic components (lysosome) in the fibroblast cells compared to M. musculus (A).
Genetics of axon damage response and axon regeneration.
The function of the nervous system relies on the transmission of information throughout an organism via projections of neuronal cells. A subset of these projections called axons relay information from one cell to another downstream cell, sometimes over long biological distances. Axons can be severed by traumatic injury; can degrade as part of the etiology of stroke or neurodegenerative diseases; and can be damaged by chemotherapeutic drugs during cancer treatment. Loss of axons also occurs during normal aging, in the absence of disease altogether. Damage to axons can result in aberrant signaling, or a complete loss of signaling if the axon is severed and fails to reconnect. Axon regeneration in the central nervous system (CNS) of mammals is extremely limited, therefore damage to the CNS often results in permanent dysfunction. Currently, there are no approved therapies to regrow damaged CNS axons in humans.
Interestingly, a previous study has demonstrated the remarkable ability of a Southeast Asian subspecies of mouse (Mus musculus castaneus) to regrow CNS axons after injury compared to other mice. We are using this natural variation in axon regeneration phenotypes between the M. m. castaneous and the more commonly used M. m. musculus to study the biology of axon damage response and regeneration.