[Inspirational text will appear here]

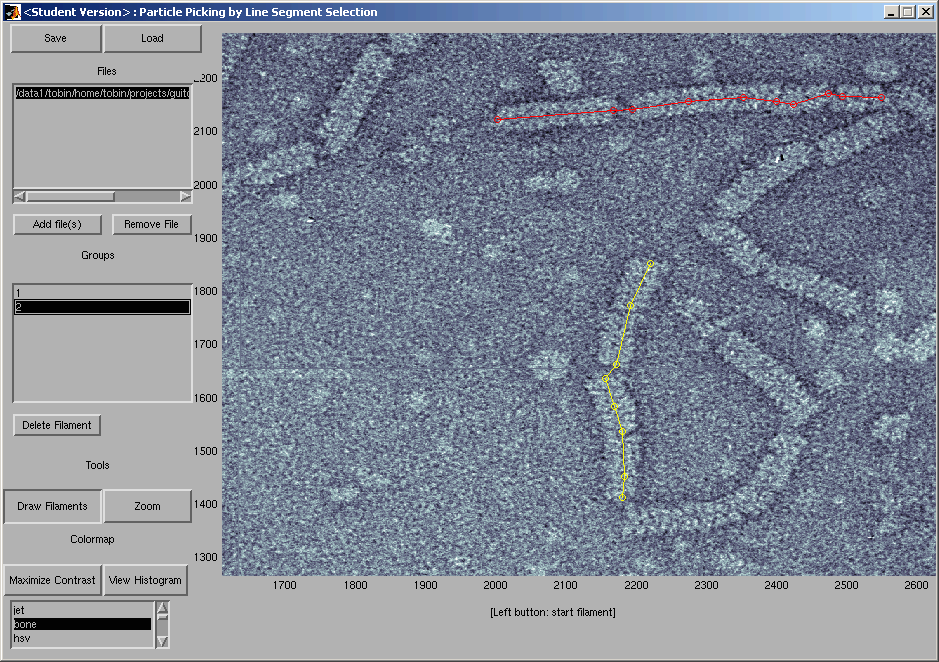
Caption: tracing helical segments of the Agrobacterium Tumefaciens T-Complex.
This software will run anywhere that MATLAB is available. You will need the `mex compiler' to compile spi_read (to read SPIDER files into Matlab), although you can download binaries for x86 linux from the spi_file web page.
If you find this software useful or interesting, I would appreciate it if you could send me a short email. My general address is tobin@splorg.org.
[nonexistant at this point]
The entire state of your selections within the Filament Picker is stored in a variable called `state', a complex object (`struct') that keeps track of the files you have loaded, the filaments (`groups') you have defined for each file, and the verticies of each filament. When you use the 'Save' button in the Filament Picker, the state object is written to a Matlab (.mat) file of your choice. In the following example the filename is "state.mat".
>> load('state.mat')
>> who
Your variables are:
ans state
>> state
state =
files: [1x2 struct]
currentfile: 2
lastgroup: 5
>> state_display(state)
+----File /home/tobin/projects/guitool/data/uacH8sptl_001.spi
| Group 1 (6 verticies)
| Group 2 (17 verticies)
| Group 3 (3 verticies)
| Group 4 (5 verticies)
+----File /home/tobin/projects/guitool/data/uacH8sptl_002.spi
| Group 5 (0 verticies)
This program was developed during summer 2002 while I was student at the Karyn Kupcinet International Science School at the Weizmann Institute of Science in Rehovot, Israel.
Copyright © 2003 by Tobin Fricke <TOBIN@SPLORG.ORG>