SIEBEL STEM CELL INSTITUTE
The Siebel Stem Cell Institute is an interactive joint program between the Berkeley Stem Cell Center and
the Stanford Institute for Stem Cell Biology and Regenerative Medicine. The Institute’s objective is to
catalyze cooperative research activities in stem cell biology at these universities, focused on some of the
grand challenges confronting the development of novel stem cell-based therapeutics to address unmet
medical needs. At UC Berkeley, these efforts are focused on supporting innovative exploratory projects
and postdoctoral fellows who show exceptional promise to become leaders in the field of regenerative
medicine.
2022 Pilot Projects Supported by the Siebel Stem Cell Institute
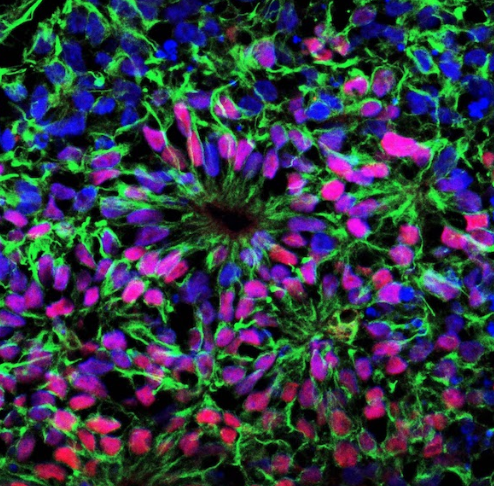
Investigating the Genetic Basis of Tuberous Sclerosis Complex
Using Human Brain Organoids and Patient Tissue
Helen Bateup Laboratory
Tuberous Sclerosis Complex (TSC) is a multi-system genetic disorder caused by mutations in the TSC1 or TSC2 genes. Along with a plethora of other clinical manifestations, TSC patients present with brain malformations called cortical tubers. Tubers are regions of the brain containing dysmorphic cells that emerge during embryonic development. Tubers disrupt the normal patterning of the cerebral cortex and contribute to a range of neurological symptoms, including epilepsy and intellectual disability. Although it is confirmed that cortical tuber regions can become epileptogenic, the exact developmental origin of the abnormal and dysmorphic cells within the tubers remains largely unknown. In this study, we will utilize recent advances in human stem cell-derived 3D cortical organoids and cortical tuber tissue samples resected from TSC patients to perform high-resolution genetic analyses. First, we will perform DNA sequencing of individual cells within the patient tuber samples to identify the genetic changes that cause cortical tubers to form.
Next, we will combine this data with single nuclei RNA sequencing of the same samples to identify alterations in gene expression within individual brain cells. Finally, we will perform single cell RNA sequencing of stem cell-derived human brain organoids with engineered TSC2 mutations to explore how closely changes seen in patient samples are reflected in cell culture models of TSC. Together this work will provide new information about the early steps of human brain development and how these may go awry in the context of TSC.
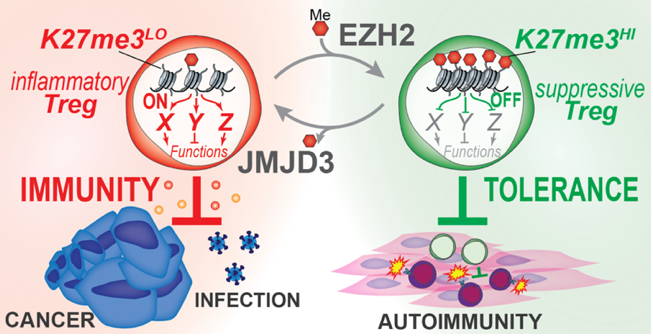
Dissecting H3K27me3-Mediated Control of Treg Plasticity in Cancer
Michel DuPage Laboratory
A fundamental question in immunology is how immune responses are controlled. Insights into mechanisms of immune regulation have led to breakthroughs in treatments for infections, cancer, and autoimmunity. Regulatory T cells (Tregs), an immunosuppressive subset of CD4+ T cells, act as gatekeepers of immune responses and are ideal therapeutic targets for modulating immune responses to cancer. However, Treg-based therapies are challenged by the central importance of Tregs in preventing autoimmunity, such that blocking Treg function systemically is highly toxic. Thus, while Tregs are heavily recruited to most tumors where they suppress antitumor immune responses, disrupting Treg function may instigate severe autoimmunity. Our objective is to discover mechanisms controlling the stem-like plasticity of Treg functions to change their activities selectively in cancer tissues from immunosuppressive to immunostimulatory. We hypothesize that Tregs uniquely adapt to cancerous tissue microenvironments by controlling chromatin structure. We propose to investigate this hypothesis by studying a single chromatin modification in Tregs, tri-methylated lysine 27 of histone H3 (H3K27me3), which appears to control Treg frequency and function specifically in organ tissues. Using gene expression analysis of wild-type, Ezh2 methylransferase-deficient, and Jmjd3 demethylase-deficient Tregs generated in our lab, we have identified a list of candidate H3K27me3-regulated genes in Tregs. We propose to use CRISPR-based gene editing in a pooled screening approach in vitro and in vivo to test the activity of candidate genes in controlling Treg stem-like plasticity.
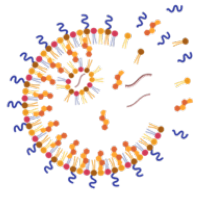
Development of Novel Solid Lipid Nanoparticles
to Improve Genome Editing in Cardiac Muscle
Kevin Healy Laboratory
Advances in genetics have revealed the molecular mechanisms that underlie many forms of cardiac disease, yet cures remain elusive. Although cardiovascular disease is the leading cause of death in the U.S, innovation in heart failure therapeutics has been sparse. Gene therapy using the CRISPR/Cas9 tool is a promising modality for the treatment of inherited and acquired cardiovascular diseases. However, genome editing in the heart suffers from low transfection efficiency of non-proliferative cardiomyocytes and diffusional barriers posed by the extracellular matrix of 3D cardiac muscle. Non-viral vectors like solid lipid nanoparticles (LNPs) hold great promise in overcoming these limitations and can make a significant breakthrough in genetic medicine due to the transient nature of mRNA transfection. This project aims to identify a solid LNP formulation that can diffuse within dense human cardiac muscle, transfect cardiomyocytes with Cas9 mRNA and gRNA, and edit the genome in a more efficient manner than current strategies.
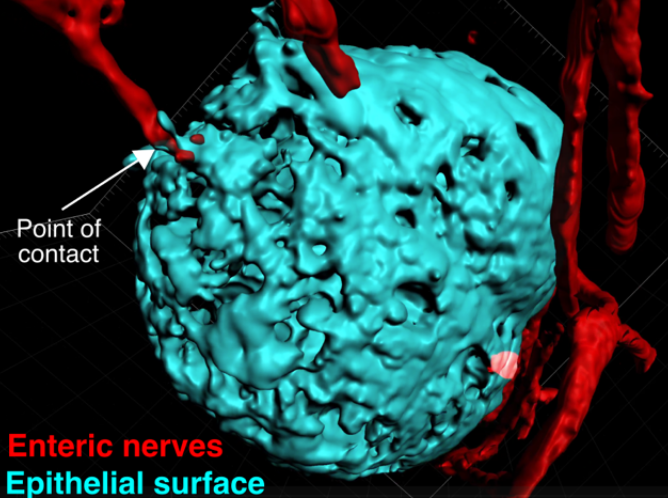
Modeling Colorectal Cancer Metastasis Using Adult Stem Cell-Derived Organoids
Megan Martik Laboratory
Metastatic cancer accounts for over 90% of the deaths from solid tumors, but few treatment options outside of traditional chemotherapy remain available to patients. Colorectal cancer represents a significant health burden, with more than one million people diagnosed with colorectal cancer every year. Once the cancer spreads to the liver and/or distant organs, the five-year survival rate drops to as low as 5%. Therefore, there is an urgent need to better understand this phenomenon and identify potential biomarkers and targeted treatment options. Over the past decade, it has become possible to expand and maintain primary epithelial stem cells in a three-dimensional culture system. This organoid technology results in self-organizing tissues that encapsulate the anatomy and cell types of the organ from which they are derived. Our proposed work will harness the power of culturing organoids from intestinal stem cells and will consequently serve as a uniquely informative model for testing colorectal cancer-causing mutations and responses to candidate drugs.
Accelerated Aging in Cerebral Organoids
Lydia Sohn Laboratory
The Census Bureau predicts that one in four Americans will be over the age of 65 by 2060. As the elderly population increases, there is an urgent need to develop preventive or therapeutic treatments for age-related neurodegeneration and cognitive decline. Few or no effective cures are available for age-associated neurodegenerative diseases, mainly because current model systems are so far removed that treatments developed in vitro fail when translated in vivo. Brain organoids, developed from human pluripotent stem cells, are three-dimensional (3D) in vitro models that have the potential to fulfill this need. Remarkably, brain organoids recapitulate aspects of the 3D tissue architecture, cellular composition, function, and gene expression profiles of the developing human brain. Still, a critical limitation of using brain organoids to study aging mechanisms and to model neurodegenerative disease is that the neurons/tissue are immature, representing those of a fetal brain. To expand the reach of organoid technologies, we propose to develop cerebral organoids that undergo accelerating aging. In this study, we will add external factors during organoid growth to accelerate maturation and create age-associated damage. We will also develop an analysis pipeline that includes RNASeq to validate our aged organoid model. Our success will lead to a platform for researchers to investigate the real-time development and progression of age-associated neurodegenerative diseases such as Alzheimer’s, Parkinson’s, or Lou Gehrig’s disease.
Using Stem Cell-Based Fat-on-a-Chip Devices to Predict Diabetes Risk
Andreas Stahl Laboratory
Type-2 diabetes mellitus remains one of the most common chronic diseases, affecting one in ten of the general US population while more than 1/3 of all US adults display signs of prediabetes characterized by impaired responses to insulin. While in general a genetic contribution to diabetes risk has been well established, it remains much less clear which genes are responsible. As a consequence, predicting diabetic risk on an individual basis, and thus determining the best intervention strategies, has not been possible. We propose to overcome this challenge by developing functional assays for insulin sensitivity using human induced pluripotent stem cells (iPSC). We will focus on the initial stages of prediabetes, i.e. adipocyte dysfunction and inflammation, by deriving fat cells (iADIPO) and macrophages (iMAC) from iPSC lines established from patients with no or strong familiar histories of diabetes. Goals will be to
generate functional iADIPO and iMAC from at least 4 high risk and 4 low risk patients, to assay iADIPO insulin sensitivity with three different assays utilizing innovative microphsyiological devices (aka organ-on-a-chip systems), and to determine the impact of introducing isogenic iMACs into the fat-on-a-chip device. Successful completion of these aims will demonstrate the feasibility of expanding this approach to a large number of human iPSC lines to enable discovery of underlying genetic features and to power novel early detection methods for the identification of high-risk individuals.
2020-21 Fellow of the Siebel Stem Cell Institute
Andrew Modzelewski, PhD

Nearly half of all mammalian genomes originate from ancient retroviral integrations. While silenced in nearly all cells, retrotransposon reactivation is a well-known phenomenon in preimplantation embryos. Many retrotransposons retain regulatory and structural features, with rare reports of influencing nearby genes. Interestingly, a subset of these splice with nearby protein coding genes, generating “chimeric transcripts” that form hundreds of novel embryonic promoters, exons and polyadenylation sites. A highly efficient electroporation based CRISPR embryo editing method I developed in the lab was used to generate retrotransposon deletions diving 5 chimeric transcripts. Disruptions led to arrested global protein synthesis, cell fate specification errors, stress induced arrest, fertilization failure, embryonic lethality and improper implantation that resembles human pregnancy complications. This project adapts proteomics, genetics, bioinformatics and CRISPR/Cas9 editing to reveal this overlooked and essential form of retrotransposons-based regulation in development and disease.
2020-21 Postdoctoral Scholars of the Siebel Stem Cell Institute
Benjamin Fellows, PhD

My research uses high-resolution Magnetic Particle Imaging (MPI) to track and monitor with single-cell sensitivity Cas9 edited hematopoietic stem cell engraftments for sickle cell disease. Already clinical trials are underway using these engraftments to cure sickle cell disease. Although promising, the engraftment treatments require additional validation of both their safety and efficacy. MPI is the ideal imaging modality to investigate hematopoietic stem cell engraftments because the tracer stays bright indefinitely within the body, and each cell provides an identical signal anywhere in the body-with zero depth attenuation. To further improve on MPI’s stem cell tracking capabilities I am developing novel high resolution superferromagnetic nanotracers that will soon provide tenfold boosts to both sensitivity and spatial resolution. These tracers will allow graft biodistribution to be monitored for months in vivo on the single cell level, a breakthrough for the entire field of stem cell tracking.
Jake Flood, PhD

A newly developed tool in our lab, EvolvR, makes use of CRISPR-Cas9 for targeted genomic mutagenesis. My research interests lie in gaining a better understanding of how EvolvR interacts with host DNA repair factors, to develop and improve tools to enable directed evolution and cell lineage tracing. Lineage tracing has been a useful strategy for investigating embryogenesis, organogenesis, tumorigenesis, and stem cell differentiation. There is still a pressing need for methods that establish connections between cell lineage and spatial patterning, and the generation of tools capable of continuous barcode diversification. I aim to apply lineage tracing to cerebral organoids, a promising model for investigating human brain development and how various disease states arise, to better understand the development and role of clonal dominance in organoid systems.
Hanqin Li, PhD

Millions of genetic variants have been identified in the human population and many of them are predicted to be associated with human diseases or evolution. However, the exact biological functions of most of those mutations are largely unknown. Towards a better understanding of human functional genomics, we use high-throughput CRISPR-based mutagenesis to introduce panels of mutations in an authentic in vitro human cell system, human pluripotent stem cells (hPSCs) and deploy functional screening of these cells and their differentiated derivatives. Our goal is to evaluate the biological impact of mutations on both molecular and physiological levels to gain knowledge of the genetic basis of a variety of diseases, including cancer, neural degenerative diseases etc. and also develop potential therapeutics.
Lin Qi, PhD

Obesity remains widespread worldwide, with over 2.5 billion adults being either overweight or obese, which dramatically increases the risk and severity of diseases ranging from type-2 diabetes mellitus, steatohepatitis, nephropathies, and even COVID-19. However, differentiation of fat-storing white adipose tissue (WAT) that shows static expression of certain hallmark genes and displays the full gamut of dynamic physiological responses as mature in vivo WAT remains a central problem in stem cell research. My research aims to create a hormonally responsive WAT derived from human induced pluripotent stem cells in a dedicated micro-physiological system. It will be an innovative platform for exploring new regulators of insulin sensitivity and metabolic mechanisms. Furthermore, it will open a door to creating an isogenic whole endocrine response system for pharmacological interrogation of normal and pathological tissue interactions.
Nike Walther, PhD
A loss of transcription control and concomitant aberrant gene expression are hallmarks of cancer and developmental disorders. Changes in gene expression occur throughout development and require precise regulation by transcription factors and gene regulatory elements to ensure the formation of healthy tissues. However, it remains poorly understood how gradual gene expression changes during stem cell differentiation are controlled by the three-dimensional genome organization and the action of transcription factors depending on the tissue context. In the rapidly self-renewing mammalian intestinal epithelium, a directional movement of differentiating intestinal stem cells underlies their maturation to differentiated cells. Using intestinal organoids that recapitulate this behavior in vitro, I am taking a multi-pronged approach combining cutting-edge genomics with state-of-the-art quantitative live imaging methods to dissect the dynamic interplay between the 3D genome organization, cell fate-determining transcription factors and target gene expression during stem cell differentiation from the tissue to the single-molecule level.